The unifying objectives of the team are:
- Test tumor growth scenarios using simple models
- Predict the future evolution of tumors
- Study and predict the effect of treatments on tumors
- Link tumor growth studied at the micro (biology) and macro (medicine) scales
- To develop image analysis tools to assist in medical diagnosis
These objectives can be divided into three research areas:
1) Modeling of clinical and biological data of gliomas.
This modeling theme consists of developing simple models, with a minimum number of parameters, and comparing them with clinical and/or biological data.
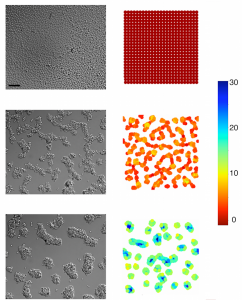
Formation of tumor cell aggregates (F98) on PEG-PLL gels, at t=0 (top), t=6h (middle) and t=12h (bottom); experiments (left), numerical simulations (right). (from Adenis L, Gontran E, Deroulers C, Grammaticos B, Juchaux M, Seksek O, Badoual M. (2020), Experimental and modeling study of the formation of cell aggregates with differential substrate adhesion, PLoS One, 15, e0222371.)
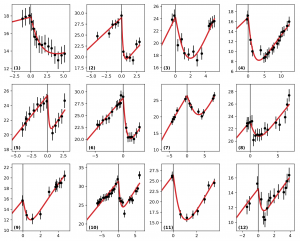
Example of automatic fitting of clinical data of tumor radius (in mm, ordinate) versus time (in years, abscissa) (from: Adenis L, Plaszczynski S, Grammaticos B, Pallud J, Badoual M, (2021), The Effect of Radiotherapy on Diffuse Low-Grade Gliomas Evolution: Confronting Theory with Clinical Data, J Pers Med, 11, 818).
Collaborations: Sainte-Anne Hospital, Institut Curie, TIMC Laboratory (Grenoble).
2) Statistical physics for gliomas.
This theme consists in studying the collective behavior of cell assemblies, based on simple rules of cell movement and cell-cell interactions and using statistical physics tools (active matter).
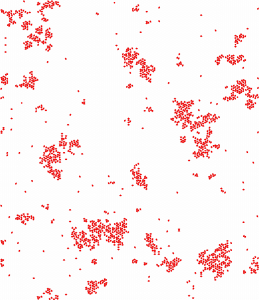
Cluster formation in polarized cell migration (from Nakamura G, Badoual M, Fabiani E and Deroulers D, (2021), Dispersal and organization of polarized cells: nonlinear diffusion and cluster formation without adhesion, J. Stat. Mech. 093501).
3) Big data analysis of histological slides.
This theme consists of data analysis, in order to feed mathematical models in the medium term, of images from the digitization of histological slides. This involves the development of software (for valorization and/or publication) for the manipulation and analysis of huge images (gigabytes).
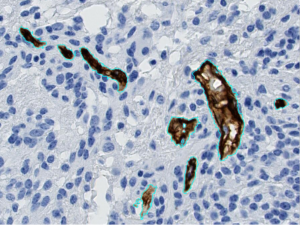
Example of automatic segmentation of blood vessels (in brown, circled in cyan blue), on a tissue sample from a glioma.
Collaborations: Sainte-Anne, Necker, Kremlin-Bicêtre hospitals and University of Caen.






